HANSOLL NA
Histology-Specific Predictors of Resistance to Neoadjuvant Chemoimmunotherapy in NSCLC
You can explore details about this study in the online abstract (P2.06.48) and the poster presented at 2025 WCLC.
Since mid-2024, I have been contributing to a prospective clinical study at Yonsei University College of Medicine focused on uncovering the genomic and immune predictors of treatment resistance in non-small cell lung cancer (NSCLC), particularly in the context of neoadjuvant chemoimmunotherapy (nCIT).
nCIT is reshaping how we treat early-stage NSCLC, yet predicting who will actually respond remains an unsolved challenge. Despite promising overall outcomes, a subset of patients experiences minimal tumor regression despite receiving PD-(L)1 blockade and chemotherapy. It was hypothesized that resistance mechanisms are not universal, but rather shaped by tumor histology—adenocarcinoma (LUAD) versus squamous cell carcinoma (LUSC).

Histology-specific predictors of neoadjuvant chemoimmunotherapy resistance in NSCLC
To explore these resistance mechanisms, I analyzed paired genomic and immune features from 72 pre-treatment tumor samples (stage I–III NSCLC). By integrating next-generation sequencing and AI-based tumor-infiltrating lymphocyte (TIL) quantification , I evaluated somatic mutations, copy-number variations (CNVs), and immune phenotypes.
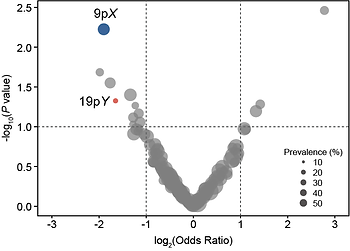
Among NSCLC patients, 36% met resistance criteria—defined as either no radiologic tumor shrinkage or ≥60% residual viable tumor after surgery. Genome-wide analysis revealed recurrent CNV loss events associated with resistance. Notably, loss of cytobands 9pX and 19pY was enriched in non-responders.

Histology-aware associations with CNVs and immune profiles
I then examined how these CNVs influence major pathologic response (MPR) and resistance. Loss of 9pX correlated with a non-inflamed immune phenotypes, suggesting that focal genomic alterations may contribute to resistance by shaping an immune-cold tumor microenvironment. Both 9pX and 19pY deletions significantly lowered MPR rates and increased resistance.
Histology stratification revealed that 9pX loss had a broad impact across all NSCLC subtypes, while 19pY loss showed a stronger effect in LUAD specifically. These associations were absent in squamous histology. Together, these findings suggest that CNVs may serve as early biomarkers for resistance, underscoring the importance of histology-aware, biomarker-driven therapeutic strategies in NSCLC.

Predictive modeling using clinico-genomic features
To evaluate the predictive value of these features, I built a logistic regression model. Clinical variables alone—such as stage or tumor proportion score—were insufficient to accurately predict response, and adding immunophenotypes offered only marginal, non-significant improvement. However, incorporating genomic alterations into the model significantly boosted predictive performance (AUC: 0.87 vs. 0.67, p=0.007).
In LUAD-specific analysis, the improvement was even more pronounced, though limited by the small sample size. This project is ongoing, with additional samples being collected to increase statistical power and further validate the model.
This study highlights histology-specific associations between genomic alterations, immune contexture, and pathologic response. CNVs—particularly 9pX and 19pY—emerged as candidate predictors in a histology-dependent manner. These findings support precision-based patient stratification to identify individuals less likely to benefit from nCIT.
I find it deeply rewarding to interpret and understand resistance from both biological and clinical lenses—linking what we see in the genome to what we observe at the bedside. This experience has strengthened my foundation in translational oncology and deepened my insight into how research can meet clinical need.